HPC User Report from L. Folle and K. Tkotz (Chair of Computer Science 5, Pattern Recognition)
Accelerating CEST MRI using neural networks
In this project we were able to take the first steps towards accelerating the acquisition of a special contrast for magnetic resonance imaging. Using neural networks certain information usually acquired can be skipped which results in an accelerated acquisition time. The resulting degraded image quality can be greatly improved by training a neural network. Thanks to the computation support of the Erlangen National High Performance Computing Center we could increase the time of the neural network training drastically.
Motivation and problem definition
Magnetic resonance imaging (MRI) allows the visualization of the anatomy of the human body with a high resolution. To further visualize biological processes such as inflammation, the way in which MR images are acquired can be adopted. Chemical exchange saturation transfer (CEST) allows the visualization of such processes and has the potential to detect the onset of rheumatic diseases. One limitation of CEST MRI to this date is the limited resolution of the acquired images, which makes the correlation with very small anatomical structures for example the lining of the knee bones impossible.
In the project we investigated the resolution limits of CEST MRI with neural networks.
Methods and codes
Building up on the influential works of Hammernik et al. and Sriram et al. we adopted the End-to-end variational network from 2D to 4D to be able to process the CEST MRI. Further, we made use of the publicly available fastMRI dataset to train the neural network. Thanks to the outstandingly well documented code repository from facebook research the code changes necessary for this project were very small and can be shared upon request.

Results
This project is still ongoing and the final results will be published at a later point in time. Some preliminary results are shown here. Using the undersampled k-space with the mask shown in Figure 1, the network is able to reconstruct the knee seen in Figure 2 which visually is close the the fully-sampled data in Figure 3. The quantitative evaluation of this project is still ongoing.
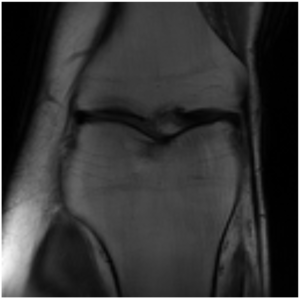
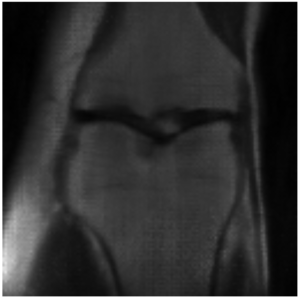
Regarding the computational efficiency of the project we could see a near linear scaling of number of GPUs utilized to reduced training time.
Outreach
We were able to publish an abstract of the presented work at the ISMRM 2022 CRIS.
Researcher’s Bio and Affiliation
Lukas Folle is a PhD candidate at the pattern recognition lab of the FAU collaborating with the rheumatology department of the University clinic in Erlangen. His work mostly focuses on bringing modern pattern recognition systems such as neural networks to the field of rheumatology.
Katharina Tkotz is a PhD candidate at the radiology department of the university clinic in Erlangen. Her work comprises of advancing MR systems to allow the visualization of metabolic processes in human bodies with a special focus on rheumatic diseases.
