HPC User Report from R. Warnock (Professorship of Systems Paleobiology)
Putting the F in FBD analyses: tree constraints or morphological data?
The fossilized birth-death (FBD) process is a model that allows us to estimate dated phylogenetic trees of living and fossil species. Using this approach, fossils are directly integrated as part of the tree, leading to a statistically coherent prior on divergence times in a Bayesian framework. The relationships between living species can be inferred using molecular sequences, but since fossils are typically not associated with molecular data, additional information is required to place fossils in the tree.
Motivation and problem definition
It is not known which approach to handling the phylogenetic uncertainty associated with the placement fossils will produce reliable results in FBD analyses. Here, we use a simulation approach, where the true underlying tree is known, to evaluate two different approaches to handling fossil placement: (1) using total-evidence analyses, which use a morphological data matrix for both fossils and living species, in addition to the molecular alignment for living species only, or (2) using topological constraints, where the user specifies monophyletic clades based on established taxonomy. We assess the impact of introducing errors into sets of constraints. In addition, we explore how variation in rates of fossil recovery or species diversification rates impact these approaches since, in reality, both processes are distinctly non-uniform.
Methods and codes
We simulated datasets of trees, fossils, morphological characters, and molecular sequences using R. We use the package FossilSim, developed by the authors of this study, to simulate fossils. To infer phylogenetic trees under the FBD process we use the Bayesian phylogenetic software BEAST2, written in the java programming language.
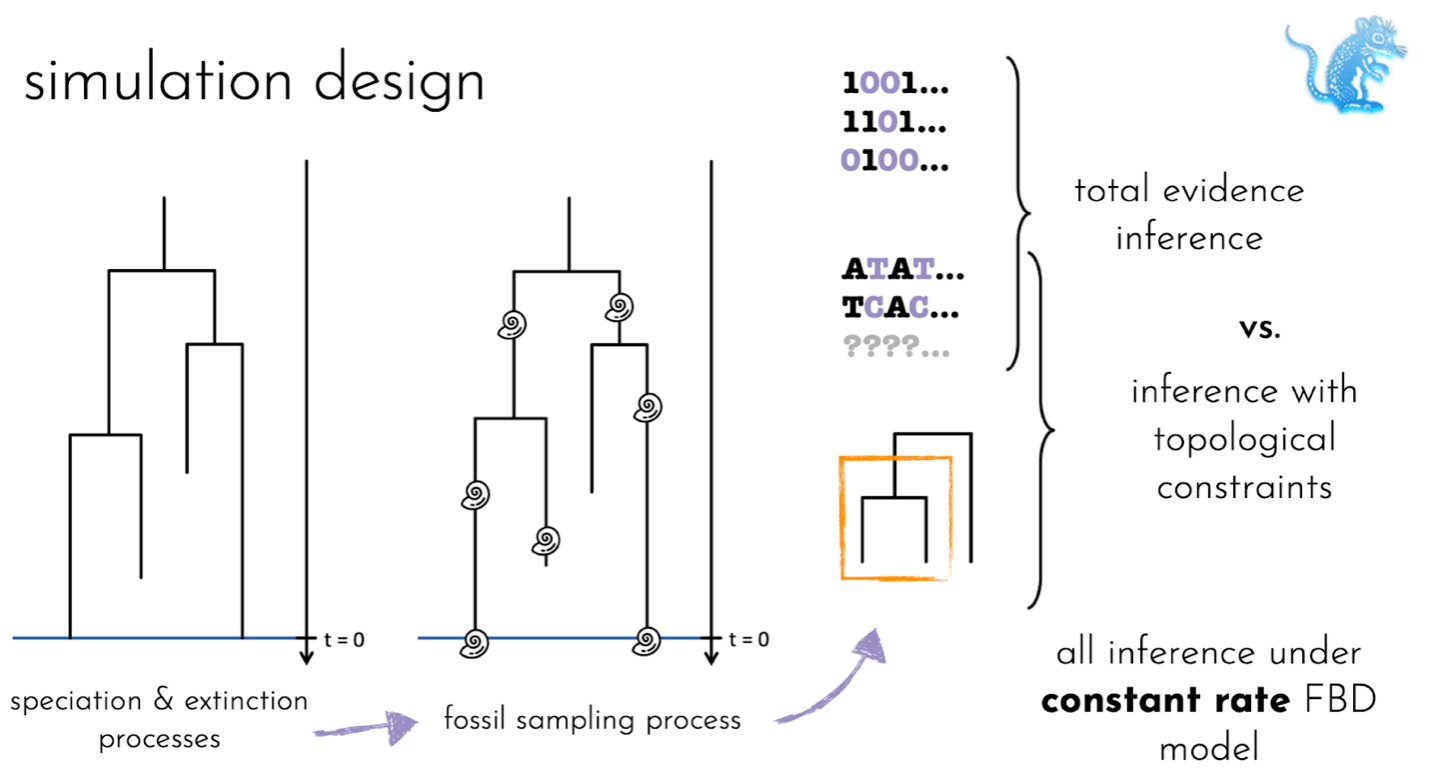
Results
We find that the tree topology is well recovered under all methods of fossil placement. Divergence times are similarly well recovered across all methods, with the exception of constraints that contain errors. We see similar patterns in datasets that include rate variation, however, relative errors in extant divergence times increase when more variation is included in the dataset, for all approaches using topological constraints, and particularly for constraints with errors. Finally, we show that trees recovered under the FBD model are more accurate than those estimated using non-time calibrated inference. Overall, our results underscore the importance of core taxonomic research, including morphological data collection and species descriptions, irrespective of the approach to handling phylogenetic uncertainty using the FBD process.
References
The study is currently in review and a preprint is available here: https://www.biorxiv.org/content/10.1101/2022.07.07.499091v1
Prof. Rachel Warnock’s research focuses on the role of fossils in phylogenetics and the application of phylogenetic models in paleobiology. She has a BSc in Genetics and an MRes in Biosystematics. She completed her PhD in paleobiology at the University of Bristol in 2014, working on Bayesian approaches to estimating divergence times. She did fellowships at the Smithsonian Museum of Natural History and ETH Zurich, before becoming a professor at Friedrich-Alexander-Universität Erlangen-Nürnberg at the GZN. Recently, she has worked on extensions of the fossilized birth-death process and approaches to estimating diversification rates. She is particularly interested in the role of fossil sampling and uncertainty in phylogenetics and macroevolution.
