HPC User Report from A. Bochicchio (Professorship of Computational Biology)
Nanoscale Architectures of Biological Membranes
We study the phase behavior of multicomponent lipid bilayers as model systems of complex biological membranes. A detailed molecular description of the membranes lateral heterogeneity is fundamental for a mechanistic understanding of e.g. receptor partitioning, oligomerization, and receptor signaling.
Motivation and problem definition
Bilayers made of multiple species of lipid molecules form the underlying structure of biological membranes.
A widely accepted paradigm describes such bilayers in cells as spatial inhomogeneous, i.e. characterized by regions that differ in local composition either permanently or transiently, that are rich in cholesterol and sphingomyelin, and range from 10-200 nm in diameter. The biological relevance of such ‘nanodomains’ stems from their implications for the spatial distribution of membrane proteins, which is influenced by the local lipid environment, and which directly affects fundamental biological processes such as cell signaling. Despite decades of investigation, our current understanding of the diversity, the properties, and the functions of such nanodomains is still very limited and, in some cases, controversial. Lipid nanodomains are elusive for most of the microscopic experimental techniques due to their small size and their dynamics, yet there is also a lack of atomistic simulation models able to describe their spontaneous formation in sufficiently simple but relevant complex membranes.
In order to connect lipid structure and domain formation, we are studying model ternary and quaternary systems involving a combination of high melting saturated lipids, low melting unsaturated lipids, cholesterol and/or sphingomyelin using Molecular Dynamics (MD) simulations. In such systems, domain formation is related to the presence of co-existing phases – a so-called liquid-ordered phase (Lo) enriched in cholesterol and sphingolipids and a liquid-disordered (Ld) phase – in phase separated regimes.
Methods and codes
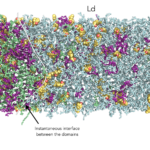
In order to cope with the involved multi-microsecond timescale for phase formation, we adopted a sequential multiscale approach combining coarse grained (CG) and atomistic (AA) MD simulations. Our approach allows to overcome the limitations on accessible time-scales and system-sizes of the AA models, and the insufficient accuracy of CG force fields. All the simulations were performed with the GROMACS software package (version 2018). While the CG MD simulations require few cores per run, the highly computational-demanding AA MD simulations are only feasible by using highly parallel computing.
Results
The chosen lipid compositions form stable coexisting liquid-ordered/liquid-disordered phases on a 10-µs time–scale at AA resolution, allowing to study the lateral organization of the lipids, and cholesterol. The molecular view that emerges is of membrane domains characterized by complex nanoscale morphology driven by phospholipid-cholesterol interactions: The Lo domains are composed by substructures of hexagonally packed saturated hydrocarbon chains nanoclusters, separated by interstitial regions enriched in cholesterol.
In the Ld domains, the low-abundant cholesterol partition to the membrane core and shows spontaneous trans-bilayer motion (flip-flop), with higher rates depending on the increasing level of lipid chains unsaturation.
The extensive AA simulations enable detailed, structure-based descriptions of relevant biological behaviors, such as protein partitioning in heterogeneous coexisting Lo/Ld domains, and cholesterol-protein interactions.
Outreach
The project was presented at the “Towards Simulating Cell Membranes: Close to Reality” workshop in Heidelberg, and at the 12th EBSA & 20th IUPAB Biophysics congress in Madrid. A publication is in preparation.
Researcher’s Bio and Affiliation
Anna Bochicchio received her Ph.D. in Computational Biophysics from RWTH Aachen University and Forschungszentrum Jülich in November 2017. In her thesis, she focused on the study of RNA-ligands and RNA-protein complexes, using atomistic MD simulations and enhanced sampling techniques. Since April 2018, she is a member of the Computational Biology group at Friedrich-Alexander Universität Erlangen-Nürnberg directed by Prof. Dr. Rainer A. Böckmann, where she is involved in the study of the phase transition in multicomponent lipid bilayers and of the lateral distribution of transmembrane peptides in bio-membrane mimetics using multi-scale molecular dynamics simulations.
